Open a SAFE product#
import xarray as xr
from xarray_eop.api import open_datatree
from xarray_eop.path import EOPath
Open a specific part of the product as a dataset#
The specific part may be determined by a given netcdf file from the SAFE or a given group corresponding to the EOPF zarr-like format.
Product stored in the local filesystem:#
import importlib.resources
SAMPLE_PATH = importlib.resources.files("tests.ut.data")
product = SAMPLE_PATH / "S3B_OL_1_ERR_sample.SEN3"
One can access the data by specifying a group corresponding to the EOPF zarr-like format:
group="/conditions/instrument"
ds = xr.open_dataset(product,file_or_group=group,engine="sentinel-3")
ds
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/xarray/namedarray/core.py:514: UserWarning: Duplicate dimension names present: dimensions {'bands'} appear more than once in dims=('bands', 'bands'). We do not yet support duplicate dimension names, but we do allow initial construction of the object. We recommend you rename the dims immediately to become distinct, as most xarray functionality is likely to fail silently if you do not. To rename the dimensions you will need to set the ``.dims`` attribute of each variable, ``e.g. var.dims=('x0', 'x1')``.
warnings.warn(
<xarray.Dataset> Size: 2kB
Dimensions: (bands: 5, detectors: 37, bands1: 5, bands2: 5)
Dimensions without coordinates: bands, detectors, bands1, bands2
Data variables:
fwhm (bands, detectors) float32 740B ...
lambda0 (bands, detectors) float32 740B ...
relative_spectral_covariance (bands1, bands2) float32 100B ...
solar_flux (bands, detectors) float32 740B ...- bands: 5
- detectors: 37
- bands1: 5
- bands2: 5
- fwhm(bands, detectors)float32...
- long_name :
- OLCI bandwidth (Full Widths at Half Maximum)
- units :
- nm
- valid_min :
- 0.0
- valid_max :
- 650.0
- ancillary_variables :
- detector_index lambda0
[185 values with dtype=float32]
- lambda0(bands, detectors)float32...
- long_name :
- OLCI characterised central wavelength
- units :
- nm
- valid_min :
- 390.0
- valid_max :
- 1040.0
- ancillary_variables :
- detector_index FWHM
[185 values with dtype=float32]
- relative_spectral_covariance(bands1, bands2)float32...
[25 values with dtype=float32]
- solar_flux(bands, detectors)float32...
- long_name :
- In-band solar irradiance, seasonally corrected
- units :
- mW.m-2.nm-1
- valid_min :
- 500.0
- valid_max :
- 2500.0
- ancillary_variables :
- detector_index lambda0
[185 values with dtype=float32]
or by opening a whole netcdf file
file="instrument_data.nc"
ds = xr.open_dataset(product,file_or_group=file,engine="sentinel-3")
ds
<xarray.Dataset> Size: 103kB
Dimensions: (rows: 103, columns: 122, bands: 5,
detectors: 37, bands1: 5, bands2: 5)
Dimensions without coordinates: rows, columns, bands, detectors, bands1, bands2
Data variables:
detector_index (rows, columns) float32 50kB ...
frame_offset (rows, columns) float32 50kB ...
lambda0 (bands, detectors) float32 740B ...
FWHM (bands, detectors) float32 740B ...
solar_flux (bands, detectors) float32 740B ...
relative_spectral_covariance (bands1, bands2) float32 100B ...
Attributes: (12/17)
netCDF_version: 4.2 of Jul 8 2020 09:28:41 $
product_name: S3B_OL_1_ERR____20230506T015316_20230506T015616_2...
title: OLCI Level 1b Product, Instrument Data Set
institution: LR1
source: IPF-OL-1-EO 06.17
history:
... ...
start_time: 2023-05-06T01:53:16.029027Z
stop_time: 2023-05-06T01:56:16.040795Z
processing_baseline: OL__L1_.003.03.00
comment:
ac_subsampling_factor: 16
al_subsampling_factor: 1- rows: 103
- columns: 122
- bands: 5
- detectors: 37
- bands1: 5
- bands2: 5
- detector_index(rows, columns)float32...
- long_name :
- Detector index
- valid_min :
- 0
- valid_max :
- 3699
[12566 values with dtype=float32]
- frame_offset(rows, columns)float32...
- long_name :
- Re-sampling along-track frame offset
- valid_min :
- -15
- valid_max :
- 15
[12566 values with dtype=float32]
- lambda0(bands, detectors)float32...
- long_name :
- OLCI characterised central wavelength
- units :
- nm
- valid_min :
- 390.0
- valid_max :
- 1040.0
- ancillary_variables :
- detector_index FWHM
[185 values with dtype=float32]
- FWHM(bands, detectors)float32...
- long_name :
- OLCI bandwidth (Full Widths at Half Maximum)
- units :
- nm
- valid_min :
- 0.0
- valid_max :
- 650.0
- ancillary_variables :
- detector_index lambda0
[185 values with dtype=float32]
- solar_flux(bands, detectors)float32...
- long_name :
- In-band solar irradiance, seasonally corrected
- units :
- mW.m-2.nm-1
- valid_min :
- 500.0
- valid_max :
- 2500.0
- ancillary_variables :
- detector_index lambda0
[185 values with dtype=float32]
- relative_spectral_covariance(bands1, bands2)float32...
[25 values with dtype=float32]
- netCDF_version :
- 4.2 of Jul 8 2020 09:28:41 $
- product_name :
- S3B_OL_1_ERR____20230506T015316_20230506T015616_20230711T065804_0179_079_117______LR1_D_NR_003.SEN3
- title :
- OLCI Level 1b Product, Instrument Data Set
- institution :
- LR1
- source :
- IPF-OL-1-EO 06.17
- history :
- references :
- S3IPF PDS 004.1 - i2r6 - Product Data Format Specification - OLCI Level 1, S3IPF PDS 002 - i1r8 - Product Data Format Specification - Product Structures, S3IPF DPM 002 - i2r9 - Detailed Processing Model - OLCI Level 1
- contact :
- s3ome@acri-st.fr
- creation_time :
- 2023-07-11T06:58:04Z
- resolution :
- [ 1080 1176 ]
- absolute_orbit_number :
- 26184
- start_time :
- 2023-05-06T01:53:16.029027Z
- stop_time :
- 2023-05-06T01:56:16.040795Z
- processing_baseline :
- OL__L1_.003.03.00
- comment :
- ac_subsampling_factor :
- 16
- al_subsampling_factor :
- 1
Product stored in S3 bucket#
It is also possible to access a product in a cloud storage. S3 cloud storage is currently the only handled object-storage.
xarray-eop provide an EOPath classe derived from pathlib.Path which supports S3 paths and well as fsspec-like URL chain.
buc_name = EOPath("s3://buc-acaw-dpr")
product = buc_name / "Samples/SAFE/S3B_OL_1_ERR____20230506T015316_20230506T015616_20230711T065804_0179_079_117______LR1_D_NR_003.SEN3"
ds = xr.open_dataset(
product,
file_or_group="instrument_data.nc",
engine="sentinel-3",
)
ds
<xarray.Dataset> Size: 11MB
Dimensions: (rows: 1023, columns: 1217, bands: 21,
detectors: 3700, bands1: 21, bands2: 21)
Dimensions without coordinates: rows, columns, bands, detectors, bands1, bands2
Data variables:
detector_index (rows, columns) float32 5MB ...
frame_offset (rows, columns) float32 5MB ...
lambda0 (bands, detectors) float32 311kB ...
FWHM (bands, detectors) float32 311kB ...
solar_flux (bands, detectors) float32 311kB ...
relative_spectral_covariance (bands1, bands2) float32 2kB ...
Attributes: (12/17)
netCDF_version: 4.2 of Jul 8 2020 09:28:41 $
product_name: S3B_OL_1_ERR____20230506T015316_20230506T015616_2...
title: OLCI Level 1b Product, Instrument Data Set
institution: LR1
source: IPF-OL-1-EO 06.17
history:
... ...
start_time: 2023-05-06T01:53:16.029027Z
stop_time: 2023-05-06T01:56:16.040795Z
processing_baseline: OL__L1_.003.03.00
comment:
ac_subsampling_factor: 16
al_subsampling_factor: 1- rows: 1023
- columns: 1217
- bands: 21
- detectors: 3700
- bands1: 21
- bands2: 21
- detector_index(rows, columns)float32...
- long_name :
- Detector index
- valid_min :
- 0
- valid_max :
- 3699
[1244991 values with dtype=float32]
- frame_offset(rows, columns)float32...
- long_name :
- Re-sampling along-track frame offset
- valid_min :
- -15
- valid_max :
- 15
[1244991 values with dtype=float32]
- lambda0(bands, detectors)float32...
- long_name :
- OLCI characterised central wavelength
- units :
- nm
- valid_min :
- 390.0
- valid_max :
- 1040.0
- ancillary_variables :
- detector_index FWHM
[77700 values with dtype=float32]
- FWHM(bands, detectors)float32...
- long_name :
- OLCI bandwidth (Full Widths at Half Maximum)
- units :
- nm
- valid_min :
- 0.0
- valid_max :
- 650.0
- ancillary_variables :
- detector_index lambda0
[77700 values with dtype=float32]
- solar_flux(bands, detectors)float32...
- long_name :
- In-band solar irradiance, seasonally corrected
- units :
- mW.m-2.nm-1
- valid_min :
- 500.0
- valid_max :
- 2500.0
- ancillary_variables :
- detector_index lambda0
[77700 values with dtype=float32]
- relative_spectral_covariance(bands1, bands2)float32...
[441 values with dtype=float32]
- netCDF_version :
- 4.2 of Jul 8 2020 09:28:41 $
- product_name :
- S3B_OL_1_ERR____20230506T015316_20230506T015616_20230711T065804_0179_079_117______LR1_D_NR_003.SEN3
- title :
- OLCI Level 1b Product, Instrument Data Set
- institution :
- LR1
- source :
- IPF-OL-1-EO 06.17
- history :
- references :
- S3IPF PDS 004.1 - i2r6 - Product Data Format Specification - OLCI Level 1, S3IPF PDS 002 - i1r8 - Product Data Format Specification - Product Structures, S3IPF DPM 002 - i2r9 - Detailed Processing Model - OLCI Level 1
- contact :
- s3ome@acri-st.fr
- creation_time :
- 2023-07-11T06:58:04Z
- resolution :
- [ 1080 1176 ]
- absolute_orbit_number :
- 26184
- start_time :
- 2023-05-06T01:53:16.029027Z
- stop_time :
- 2023-05-06T01:56:16.040795Z
- processing_baseline :
- OL__L1_.003.03.00
- comment :
- ac_subsampling_factor :
- 16
- al_subsampling_factor :
- 1
Open the whole SAFE product as a datatree structure#
dt = open_datatree(product)
dt
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - time_stamp(rows)datetime64[ns]dask.array<chunksize=(1023,), meta=np.ndarray>
- standard_name :
- time
- long_name :
- Elapsed time since 01 Jan 2000 0h
Array Chunk Bytes 7.99 kiB 7.99 kiB Shape (1023,) (1023,) Dask graph 1 chunks in 1 graph layer Data type datetime64[ns] numpy.ndarray
- oa01_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa01
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa01_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa02_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa02
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa02_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa03_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa03
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa03_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa04_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa04
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa04_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa05_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa05
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa05_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa06_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa06
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa06_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa07_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa07
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa07_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa08_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa08
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa08_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa09_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa09
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa09_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa10_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa10
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa10_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa11_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa11
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa11_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa12_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa12
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa12_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa13_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa13
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa13_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa14_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa14
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa14_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa15_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa15
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa15_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa16_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa16
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa16_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa17_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa17
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa17_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa18_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa18
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa18_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa19_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa19
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa19_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa20_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa20
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa20_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa21_radiance(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa21
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa21_radiance_unc
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray
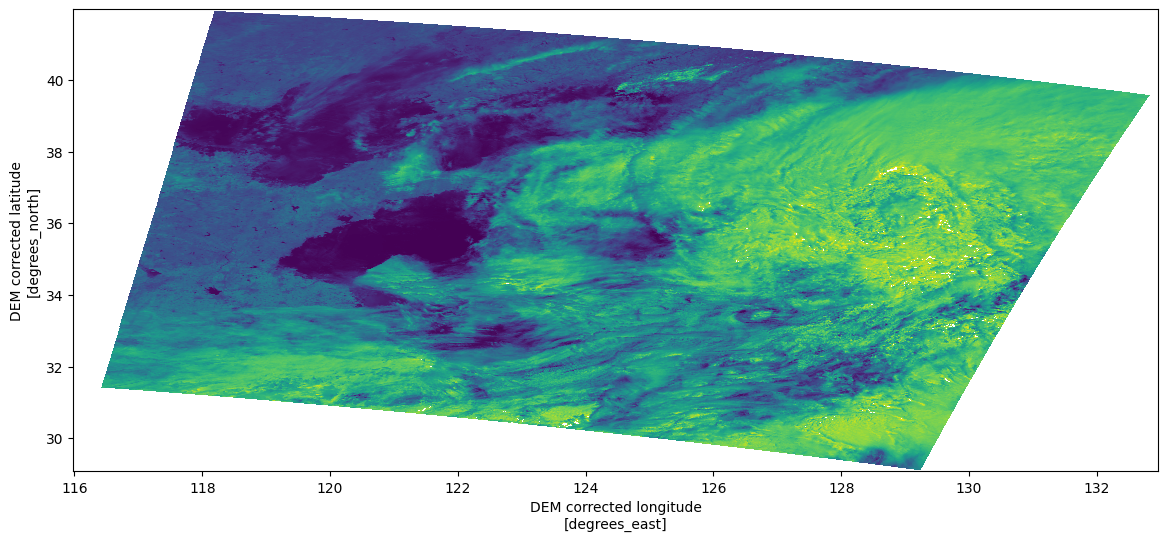
<xarray.DatasetView> Size: 125MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> time_stamp (rows) datetime64[ns] 8kB dask.array<chunksize=(1023,), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: (12/21) oa01_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa02_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa03_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa04_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa05_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa06_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> ... ... oa16_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa17_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa18_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa19_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa20_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa21_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray>measurements- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray
- frame_offset(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- Re-sampling along-track frame offset
- valid_min :
- -15
- valid_max :
- 15
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - detector_index(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- Detector index
- valid_min :
- 0
- valid_max :
- 3699
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - altitude(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- altitude
- long_name :
- DEM corrected altitude
- units :
- m
- valid_min :
- -1000
- valid_max :
- 9000
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 35MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: frame_offset (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> detector_index (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> altitude (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray>image- tie_rows: 1023
- tie_columns: 77
- latitude(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - longitude(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray
- oaa(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Observation (Viewing) Azimuth Angle
- units :
- degrees
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - oza(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Observation (Viewing) Zenith Angle
- units :
- degrees
- valid_min :
- 0
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - saa(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Sun Azimuth Angle
- units :
- degrees
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - sza(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Sun Zenith Angle
- units :
- degrees
- valid_min :
- 0
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 4MB Dimensions: (tie_rows: 1023, tie_columns: 77) Coordinates: latitude (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> longitude (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> Dimensions without coordinates: tie_rows, tie_columns Data variables: oaa (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> oza (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> saa (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> sza (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray>geometry- tie_rows: 1023
- tie_columns: 77
- tie_pressure_levels: 25
- wind_vectors: 2
- tie_pressure_levels(tie_pressure_levels)float321e+03 950.0 925.0 ... 3.0 2.0 1.0
- standard_name :
- air_pressure
- long_name :
- Reference pressure level
- units :
- hPa
- valid_min :
- 0.0
- valid_max :
- 1500.0
array([1000., 950., 925., 900., 850., 800., 700., 600., 500., 400., 300., 250., 200., 150., 100., 70., 50., 30., 20., 10., 7., 5., 3., 2., 1.], dtype=float32)
- atmospheric_temperature_profile(tie_rows, tie_columns, tie_pressure_levels)float32dask.array<chunksize=(1023, 77, 25), meta=np.ndarray>
- standard_name :
- air_temperature
- long_name :
- Air temperature profile
- units :
- K
- valid_min :
- 0.0
- valid_max :
- 400.0
- ancillary_variables :
- reference_pressure_level
Array Chunk Bytes 7.51 MiB 7.51 MiB Shape (1023, 77, 25) (1023, 77, 25) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - horizontal_wind(tie_rows, tie_columns, wind_vectors)float32dask.array<chunksize=(1023, 77, 2), meta=np.ndarray>
- long_name :
- Horizontal wind vector at 10m altitude
- units :
- m.s-1
- valid_min :
- -100.0
- valid_max :
- 100.0
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77, 2) (1023, 77, 2) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - humidity(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- relative_humidity
- long_name :
- Relative humidity
- units :
- %
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - sea_level_pressure(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- air_pressure_at_sea_level
- long_name :
- Mean sea level pressure
- units :
- hPa
- valid_min :
- 0.0
- valid_max :
- 1500.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - total_columnar_water_vapour(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- atmosphere_water_vapor_content
- long_name :
- Total column water vapour
- units :
- kg.m-2
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - total_ozone(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- atmosphere_mass_content_of_ozone
- long_name :
- Total columnar ozone
- units :
- kg.m-2
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 10MB Dimensions: (tie_rows: 1023, tie_columns: 77, tie_pressure_levels: 25, wind_vectors: 2) Coordinates: * tie_pressure_levels (tie_pressure_levels) float32 100B 1e+03... Dimensions without coordinates: tie_rows, tie_columns, wind_vectors Data variables: atmospheric_temperature_profile (tie_rows, tie_columns, tie_pressure_levels) float32 8MB dask.array<chunksize=(1023, 77, 25), meta=np.ndarray> horizontal_wind (tie_rows, tie_columns, wind_vectors) float32 630kB dask.array<chunksize=(1023, 77, 2), meta=np.ndarray> humidity (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> sea_level_pressure (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> total_columnar_water_vapour (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> total_ozone (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray>meteorology- bands: 21
- detectors: 3700
- bands1: 21
- bands2: 21
- fwhm(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- OLCI bandwidth (Full Widths at Half Maximum)
- units :
- nm
- valid_min :
- 0.0
- valid_max :
- 650.0
- ancillary_variables :
- detector_index lambda0
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - lambda0(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- OLCI characterised central wavelength
- units :
- nm
- valid_min :
- 390.0
- valid_max :
- 1040.0
- ancillary_variables :
- detector_index FWHM
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - relative_spectral_covariance(bands1, bands2)float32dask.array<chunksize=(21, 21), meta=np.ndarray>
Array Chunk Bytes 1.72 kiB 1.72 kiB Shape (21, 21) (21, 21) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - solar_flux(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- In-band solar irradiance, seasonally corrected
- units :
- mW.m-2.nm-1
- valid_min :
- 500.0
- valid_max :
- 2500.0
- ancillary_variables :
- detector_index lambda0
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 934kB Dimensions: (bands: 21, detectors: 3700, bands1: 21, bands2: 21) Dimensions without coordinates: bands, detectors, bands1, bands2 Data variables: fwhm (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray> lambda0 (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray> relative_spectral_covariance (bands1, bands2) float32 2kB dask.array<chunksize=(21, 21), meta=np.ndarray> solar_flux (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray>instrument
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*conditions- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 9.50 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float64 numpy.ndarray
- quality_flags(rows, columns)uint32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- Classification and quality flags
- flag_masks :
- [2147483648 1073741824 536870912 268435456 134217728 67108864 33554432 16777216 8388608 4194304 2097152 1048576 524288]
- flag_meanings :
- land coastline fresh_inland_water tidal_region bright straylight_risk invalid cosmetic duplicated sun-glint_risk dubious saturated partially_saturated
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type uint32 numpy.ndarray - oa01_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa01
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa02_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa02
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa03_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa03
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa04_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa04
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa05_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa05
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa06_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa06
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa07_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa07
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa08_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa08
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa09_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa09
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa10_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa10
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa11_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa11
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa12_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa12
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa13_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa13
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa14_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa14
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa15_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa15
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa16_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa16
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa17_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa17
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa18_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa18
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa19_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa19
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa20_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa20
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - oa21_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1217), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa21
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.75 MiB Shape (1023, 1217) (1023, 1217) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 129MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: (12/22) quality_flags (rows, columns) uint32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa01_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa02_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa03_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa04_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa05_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> ... ... oa16_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa17_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa18_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa19_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa20_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray> oa21_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1217), meta=np.ndarray>quality
In the case of the Sentinel-3 product, data is a collection of netcdf files. To allow lazy loading, each variable is first computed and then rechunk. To speed up the process, it is possible to make a temporary copy of the product in the local filesystem, by setting the optional argument fs_copy to True
dt = open_datatree(product,fs_copy=True)
dt
/opt/hostedtoolcache/Python/3.11.9/x64/lib/python3.11/site-packages/xarray/namedarray/core.py:514: UserWarning: Duplicate dimension names present: dimensions {'bands'} appear more than once in dims=('bands', 'bands'). We do not yet support duplicate dimension names, but we do allow initial construction of the object. We recommend you rename the dims immediately to become distinct, as most xarray functionality is likely to fail silently if you do not. To rename the dimensions you will need to set the ``.dims`` attribute of each variable, ``e.g. var.dims=('x0', 'x1')``.
warnings.warn(
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray - time_stamp(rows)datetime64[ns]dask.array<chunksize=(1023,), meta=np.ndarray>
- standard_name :
- time
- long_name :
- Elapsed time since 01 Jan 2000 0h
Array Chunk Bytes 7.99 kiB 7.99 kiB Shape (1023,) (1023,) Dask graph 1 chunks in 2 graph layers Data type datetime64[ns] numpy.ndarray
- oa01_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa01
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa01_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa02_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa02
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa02_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa03_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa03
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa03_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa04_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa04
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa04_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa05_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa05
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa05_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa06_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa06
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa06_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa07_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa07
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa07_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa08_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa08
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa08_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa09_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa09
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa09_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa10_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa10
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa10_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa11_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa11
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa11_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa12_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa12
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa12_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa13_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa13
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa13_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa14_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa14
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa14_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa15_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa15
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa15_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa16_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa16
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa16_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa17_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa17
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa17_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa18_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa18
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa18_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa19_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa19
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa19_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa20_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa20
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa20_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa21_radiance(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- toa_upwelling_spectral_radiance
- long_name :
- TOA radiance for OLCI acquisition band Oa21
- units :
- mW.m-2.sr-1.nm-1
- valid_min :
- 0
- valid_max :
- 65534
- ancillary_variables :
- Oa21_radiance_unc
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 125MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> time_stamp (rows) datetime64[ns] 8kB dask.array<chunksize=(1023,), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: (12/21) oa01_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa02_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa03_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa04_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa05_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa06_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> ... ... oa16_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa17_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa18_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa19_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa20_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa21_radiance (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray>measurements- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray
- frame_offset(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- Re-sampling along-track frame offset
- valid_min :
- -15
- valid_max :
- 15
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - detector_index(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- Detector index
- valid_min :
- 0
- valid_max :
- 3699
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - altitude(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- altitude
- long_name :
- DEM corrected altitude
- units :
- m
- valid_min :
- -1000
- valid_max :
- 9000
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 35MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: frame_offset (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> detector_index (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> altitude (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray>image- tie_rows: 1023
- tie_columns: 77
- latitude(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - longitude(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray
- oaa(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Observation (Viewing) Azimuth Angle
- units :
- degrees
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - oza(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Observation (Viewing) Zenith Angle
- units :
- degrees
- valid_min :
- 0
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - saa(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Sun Azimuth Angle
- units :
- degrees
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - sza(tie_rows, tie_columns)float64dask.array<chunksize=(1023, 77), meta=np.ndarray>
- long_name :
- Sun Zenith Angle
- units :
- degrees
- valid_min :
- 0
- valid_max :
- 180000000
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray
<xarray.DatasetView> Size: 4MB Dimensions: (tie_rows: 1023, tie_columns: 77) Coordinates: latitude (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> longitude (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> Dimensions without coordinates: tie_rows, tie_columns Data variables: oaa (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> oza (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> saa (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray> sza (tie_rows, tie_columns) float64 630kB dask.array<chunksize=(1023, 77), meta=np.ndarray>geometry- tie_rows: 1023
- tie_columns: 77
- tie_pressure_levels: 25
- wind_vectors: 2
- tie_pressure_levels(tie_pressure_levels)float321e+03 950.0 925.0 ... 3.0 2.0 1.0
- standard_name :
- air_pressure
- long_name :
- Reference pressure level
- units :
- hPa
- valid_min :
- 0.0
- valid_max :
- 1500.0
array([1000., 950., 925., 900., 850., 800., 700., 600., 500., 400., 300., 250., 200., 150., 100., 70., 50., 30., 20., 10., 7., 5., 3., 2., 1.], dtype=float32)
- atmospheric_temperature_profile(tie_rows, tie_columns, tie_pressure_levels)float32dask.array<chunksize=(1023, 77, 25), meta=np.ndarray>
- standard_name :
- air_temperature
- long_name :
- Air temperature profile
- units :
- K
- valid_min :
- 0.0
- valid_max :
- 400.0
- ancillary_variables :
- reference_pressure_level
Array Chunk Bytes 7.51 MiB 7.51 MiB Shape (1023, 77, 25) (1023, 77, 25) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - horizontal_wind(tie_rows, tie_columns, wind_vectors)float32dask.array<chunksize=(1023, 77, 2), meta=np.ndarray>
- long_name :
- Horizontal wind vector at 10m altitude
- units :
- m.s-1
- valid_min :
- -100.0
- valid_max :
- 100.0
Array Chunk Bytes 615.40 kiB 615.40 kiB Shape (1023, 77, 2) (1023, 77, 2) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - humidity(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- relative_humidity
- long_name :
- Relative humidity
- units :
- %
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - sea_level_pressure(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- air_pressure_at_sea_level
- long_name :
- Mean sea level pressure
- units :
- hPa
- valid_min :
- 0.0
- valid_max :
- 1500.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - total_columnar_water_vapour(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- atmosphere_water_vapor_content
- long_name :
- Total column water vapour
- units :
- kg.m-2
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - total_ozone(tie_rows, tie_columns)float32dask.array<chunksize=(1023, 77), meta=np.ndarray>
- standard_name :
- atmosphere_mass_content_of_ozone
- long_name :
- Total columnar ozone
- units :
- kg.m-2
- valid_min :
- 0.0
- valid_max :
- 100.0
Array Chunk Bytes 307.70 kiB 307.70 kiB Shape (1023, 77) (1023, 77) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 10MB Dimensions: (tie_rows: 1023, tie_columns: 77, tie_pressure_levels: 25, wind_vectors: 2) Coordinates: * tie_pressure_levels (tie_pressure_levels) float32 100B 1e+03... Dimensions without coordinates: tie_rows, tie_columns, wind_vectors Data variables: atmospheric_temperature_profile (tie_rows, tie_columns, tie_pressure_levels) float32 8MB dask.array<chunksize=(1023, 77, 25), meta=np.ndarray> horizontal_wind (tie_rows, tie_columns, wind_vectors) float32 630kB dask.array<chunksize=(1023, 77, 2), meta=np.ndarray> humidity (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> sea_level_pressure (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> total_columnar_water_vapour (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray> total_ozone (tie_rows, tie_columns) float32 315kB dask.array<chunksize=(1023, 77), meta=np.ndarray>meteorology- bands: 21
- detectors: 3700
- bands1: 21
- bands2: 21
- fwhm(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- OLCI bandwidth (Full Widths at Half Maximum)
- units :
- nm
- valid_min :
- 0.0
- valid_max :
- 650.0
- ancillary_variables :
- detector_index lambda0
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - lambda0(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- OLCI characterised central wavelength
- units :
- nm
- valid_min :
- 390.0
- valid_max :
- 1040.0
- ancillary_variables :
- detector_index FWHM
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray - relative_spectral_covariance(bands1, bands2)float32dask.array<chunksize=(21, 21), meta=np.ndarray>
Array Chunk Bytes 1.72 kiB 1.72 kiB Shape (21, 21) (21, 21) Dask graph 1 chunks in 1 graph layer Data type float32 numpy.ndarray - solar_flux(bands, detectors)float32dask.array<chunksize=(21, 3700), meta=np.ndarray>
- long_name :
- In-band solar irradiance, seasonally corrected
- units :
- mW.m-2.nm-1
- valid_min :
- 500.0
- valid_max :
- 2500.0
- ancillary_variables :
- detector_index lambda0
Array Chunk Bytes 303.52 kiB 303.52 kiB Shape (21, 3700) (21, 3700) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 934kB Dimensions: (bands: 21, detectors: 3700, bands1: 21, bands2: 21) Dimensions without coordinates: bands, detectors, bands1, bands2 Data variables: fwhm (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray> lambda0 (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray> relative_spectral_covariance (bands1, bands2) float32 2kB dask.array<chunksize=(21, 21), meta=np.ndarray> solar_flux (bands, detectors) float32 311kB dask.array<chunksize=(21, 3700), meta=np.ndarray>instrument
<xarray.DatasetView> Size: 0B Dimensions: () Data variables: *empty*conditions- rows: 1023
- columns: 1217
- latitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- latitude
- long_name :
- DEM corrected latitude
- units :
- degrees_north
- valid_min :
- -90000000
- valid_max :
- 90000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray - longitude(rows, columns)float64dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- standard_name :
- longitude
- long_name :
- DEM corrected longitude
- units :
- degrees_east
- valid_min :
- -180000000
- valid_max :
- 180000000
Array Chunk Bytes 9.50 MiB 7.99 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float64 numpy.ndarray
- quality_flags(rows, columns)uint32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- Classification and quality flags
- flag_masks :
- [2147483648 1073741824 536870912 268435456 134217728 67108864 33554432 16777216 8388608 4194304 2097152 1048576 524288]
- flag_meanings :
- land coastline fresh_inland_water tidal_region bright straylight_risk invalid cosmetic duplicated sun-glint_risk dubious saturated partially_saturated
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type uint32 numpy.ndarray - oa01_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa01
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa02_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa02
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa03_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa03
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa04_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa04
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa05_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa05
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa06_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa06
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa07_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa07
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa08_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa08
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa09_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa09
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa10_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa10
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa11_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa11
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa12_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa12
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa13_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa13
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa14_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa14
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa15_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa15
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa16_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa16
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa17_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa17
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa18_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa18
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa19_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa19
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa20_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa20
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray - oa21_radiance_unc(rows, columns)float32dask.array<chunksize=(1023, 1024), meta=np.ndarray>
- long_name :
- log10 scaled Radiometric Uncertainty Estimate for OLCI acquisition band Oa21
- units :
- lg(re mW.m-2.sr-1.nm-1)
- valid_min :
- 0
- valid_max :
- 254
Array Chunk Bytes 4.75 MiB 4.00 MiB Shape (1023, 1217) (1023, 1024) Dask graph 2 chunks in 2 graph layers Data type float32 numpy.ndarray
<xarray.DatasetView> Size: 129MB Dimensions: (rows: 1023, columns: 1217) Coordinates: latitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> longitude (rows, columns) float64 10MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> Dimensions without coordinates: rows, columns Data variables: (12/22) quality_flags (rows, columns) uint32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa01_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa02_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa03_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa04_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa05_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> ... ... oa16_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa17_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa18_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa19_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa20_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray> oa21_radiance_unc (rows, columns) float32 5MB dask.array<chunksize=(1023, 1024), meta=np.ndarray>quality
dt.measurements.oa17_radiance.plot()
<matplotlib.collections.QuadMesh at 0x7f99acdb4bd0>
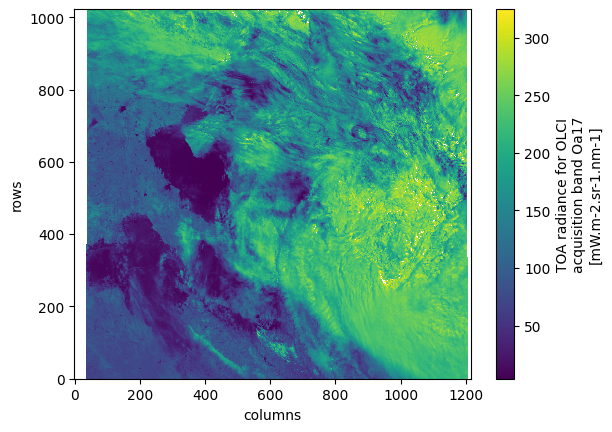
import matplotlib.pyplot as plt
plt.figure(figsize=(14, 6))
ax = plt.axes()
dt.measurements.oa17_radiance.plot.pcolormesh(
ax=ax,
x="longitude", y="latitude", add_colorbar=False
)
<matplotlib.collections.QuadMesh at 0x7f99acc4d3d0>